I recently encountered the surprisingly difficult task of processing a PDB file
in Python.
While reading fixed-width files is relatively trivial in certain old-fashioned
languages with support for complex format statements,
like my beloved Fortran,
Python makes it
less simple.
Given the myriad record types
that frequently appear in such files,
doing this processing on an ad-hoc basis seems like a terrible idea.
Thankfully,
the fields in these records lend themselves nicely to implementation as objects,
so I went ahead and did just that.
I’m by no means the first person to write a package for this purpose—searching PyPI for “pdb” turns up a lot of results, even after references to the similarly named debugger are discarded. These packages are uniformly impressive and provide powerful sets of functions for fetching, modifying, and converting PDB files. I would recommend many of them to friends and family. But that’s not what I was looking for.
I wanted a simple, lightweight package that converts PDB records into Python objects and back again, something I can drop into a script when I want to perform some arbitrary calculation of my own.
For example, I wanted to find the heavy atom closest to a known point. I’m sure many of the packages on PyPI can do that, and cpptraj almost certainly can. But the first thing that came to mind was
min(atoms,
key=lambda a: a.distance(point))
Doesn’t that look beautiful? I don’t want to install a whole package or dig through the entire AMBER manual to do something I already know how to do. Reading the data in the first place was the only hard part.
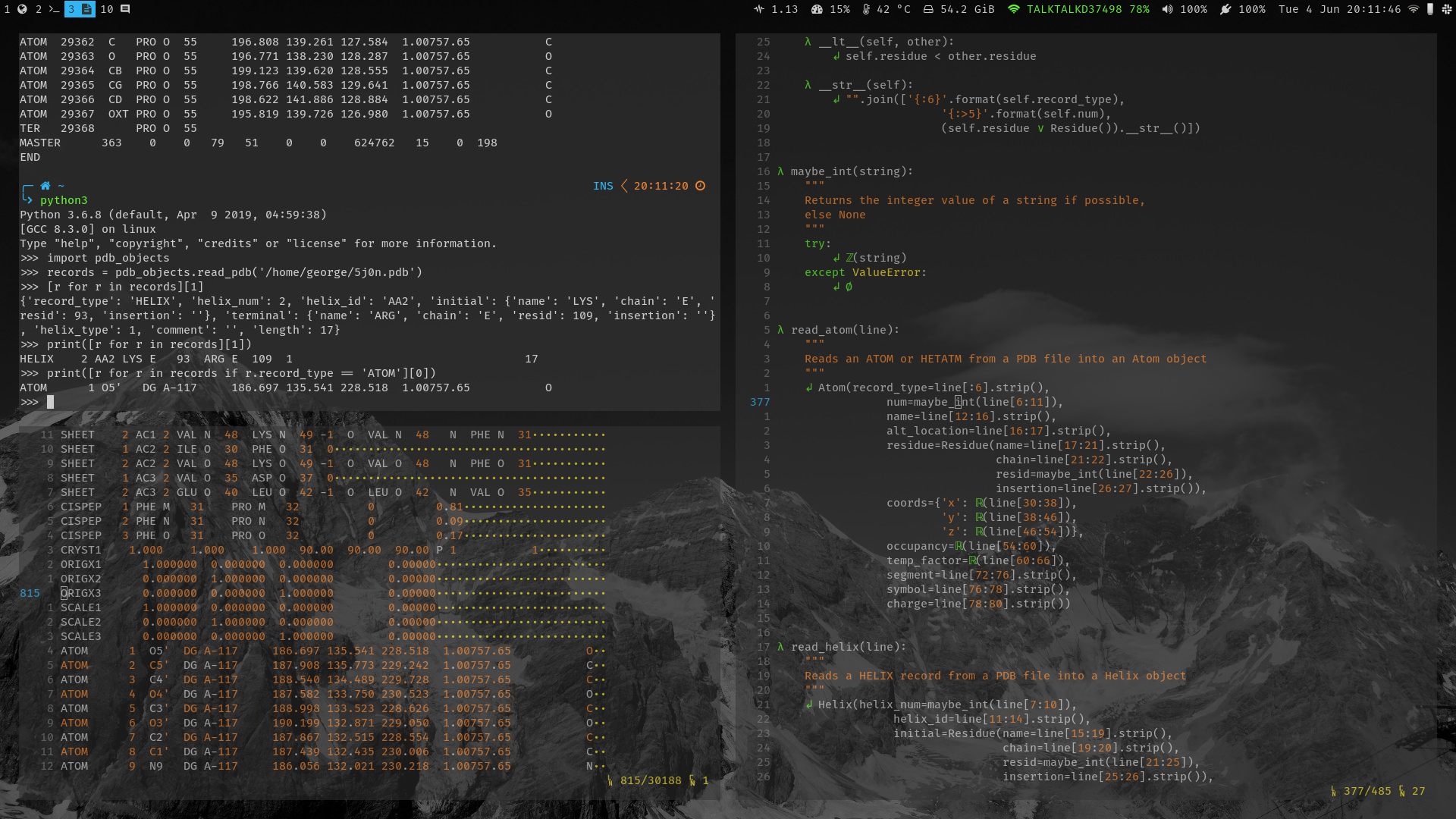
Here’s what the function to read in an ATOM record looks like:
def read_atom(line):
"""
Reads an ATOM or HETATM from a PDB file into an Atom object
"""
return Atom(record_type=line[:6].strip(),
num=maybe_int(line[6:11]),
name=line[12:16].strip(),
alt_location=line[16:17].strip(),
residue=Residue(name=line[17:21].strip(),
chain=line[21:22].strip(),
resid=maybe_int(line[22:26]),
insertion=line[26:27].strip()),
coords=Coords(x=maybe_float(line[30:38]),
y=maybe_float(line[38:46]),
z=maybe_float(line[46:54])),
occupancy=maybe_float(line[54:60]),
temp_factor=maybe_float(line[60:66]),
segment=line[72:76].strip(),
symbol=line[76:78].strip(),
charge=line[78:80].strip())
While I’m sure there are neater ways, that’s roughly as good as it gets. And if I want to manipulate the structure and write out the result, I can’t really discard very many of those fields. It’s clearly better to outsource the ugliness to a friendly package than to do that every time I want to read a PDB.
The result is an Atom object, which looks a bit like this:
{'record_type': 'ATOM',
'num': 1,
'name': "O5'",
'alt_location': '',
'residue': {'name': 'DG',
'chain': 'A',
'resid': -117,
'insertion': ''},
'coords': {'x': 186.697,
'y': 135.541,
'z': 228.518},
'occupancy': 1.0,
'temp_factor': 757.65,
'segment': '',
'symbol': 'O',
'charge': ''}
That’s much easier to work with. You may have noticed that residues and coördinates are objects too, so these elements can be easily shared across record types and a load of powerful and expressive methods can be exposed.
The package currently (v0.1.2) supports the following record types:
ATOMHETATMTERHELIXSHEET
A number of useful operators are intuitively defined.
Since PDBs are inherently ordered,
> and < do what you’d expect.
Equality is defined using the unique (in a well-formed PDB) identifiers,
so two objects representing the same atom/terminator/structural element are
equal even if their other properties are not (or undefined).
The __contains__ method is defined for residues,
so that if atom in residue is a valid construct.
Coördinates can be added and subtracted together,
and multiplied or divided by scalars.
Printing any of the objects (or otherwise casting it to a string) results in a correctly formatted PDB record like this one:
ATOM 1 O5' DG A-117 186.697 135.541 228.518 1.00757.65 O
So print(*records, sep='\n') gives you a PDB,
as long as records is a list of objects
like the one the handy read_pdb() function gives you.
This isn’t a fully-fledged PDB-manipulating package. It’s not trying to be. But it has made my life a bit easier, and maybe it will help you too.
The source code is available (under an MIT license, so you can do whatever you want with it) on GitHub, and the package is in PyPI, so you can just
$ pip3 install pdb-objects
then stick
import pdb_objects
at the top of your script and go get a coffee. You deserve it.